X-ray laser decodes “off” switch for cell signals
Findings may advance development of accurately targeted drugs
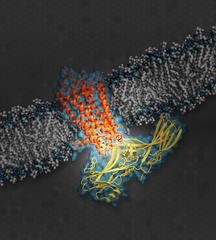
The illustration shows the signaling protein arrestin (yellow), while docked with rhodopsin (orange), a G protein-coupled receptor (GPCR). GPCRs serve an important role in a cellular signaling network. (Illustration: SLAC National Accelerator Laboratory)
Using the world's strongest X-ray laser, scientists have gained unprecedented insights into the regulation of biological cells. Like a super microscope, the Linac Coherent Light Source (LCLS) at the SLAC National Accelerator Center in the U.S. unravelled the workings of a central “off” switch in cell signalling in atomic detail. The findings can pave the way for the development of more accurately targeted drugs. The team, including researchers from CFEL, presents the work in the current issue of Nature.
Prof. Eric Xu from the Van Andel Research Institute in the U.S. and his collaborators investigated rhodopsin, the light sensitive pigment in the retina of the eye. Rhodopsin belongs to a larger class of proteins in the body. These are G protein-coupled receptors or GPCRs that are medically important since they are targeted by about a third of all prescription drugs on the market – from pain killers to high blood-pressure pills.
These receptors transmit signals via a family of proteins called guanine nucleotide-binding proteins or G proteins. In all signalling processes, such as those in vision or neural activity for example, turning off the signal is just as important as turning it on. Members of a different family of proteins, called arrestins, can block G proteins from docking to GPCR. This way, arrestins can switch off the signalling. Arrestins serve as an important “off” switch in the regulation of biological cells.
However, exactly how the arrestins bind to the receptors was unknown until now. From the data of the X-ray laser experiments, the scientists could produce the first three dimensional map of an arrestin bound to a receptor. “This study is a critical first step and provides key insight into the structural interactions in these protein complexes,” said Dr. Jeffrey L. Benovic, a biochemistry and molecular biology professor at Thomas Jefferson University in Philadelphia who specializes in such research but was not a part of this study. “This work has tremendous therapeutic implications.”
Many of the available drugs that activate or deactivate GPCRs block both G proteins and arrestins from docking. “The new paradigm in drug discovery is that you want to find this selective pathway – how to activate either the arrestin pathway or the G-protein pathway but not both – for a better effect,” said Xu. The present study might serve as a model for future investigations of other receptors sensitive to arrestins. “While this particular sample serves a specific function in the body, people may start to use this research as a model for how GPCRs, in general, can interact with signalling proteins,” said Xu.
For the investigation, the researchers had grown tiny crystals of rhodopsin with docked arrestin. The atomic structure of crystals can be determined from the way the crystals diffract X-rays. This X-ray crystallography today is a standard tool in structural biology and many other disciplines.
“Biomolecules are often quite reluctant to form a crystal, as it contradicts their natural function,” explained DESY scientist Dr. Anton Barty from CFEL, who was heavily involved in the data analysis. In the rhodopsin study, the researchers had to use microcrystals, measuring just thousandths of a millimetre. “Protein microcrystals are normally very hard to study, but thanks to the bright X-rays from the LCLS good quality diffraction patterns of some 19 000 microcrystals could be captured, and from the combination of these images the structure of the molecular complex could be determined,” said Barty.
This method of serial femtosecond X-ray crystallography was substantially developed by CUI scientist Prof. Henry Chapman and his CFEL team. “The present work shows the enormous potential of X-ray lasers for structure determination,” emphasized Chapman, head of the CFEL-DESY division for coherent imaging. This large complex was one of the most difficult structures studied to date by crystallography, further demonstrating the power of the new approach.
In addition to scientists from the Van Andel Research Institute, SLAC, and DESY, the study also included researchers from: Arizona State University, University of Southern California, National University of Singapore, New York Structural Biology Center, The Scripps Research Institute, University of California, Los Angeles, University of Toronto, Vanderbilt University, Beijing Computational Science Research Center in China, the University of Wisconsin-Milwaukee, Chinese Academy of Sciences, Paul Scherrer Institute in Switzerland, Trinity College in Ireland, University of Chicago and the University of Konstanz.